Molecular diagnosis technology uses molecular biology methods to detect the expression and structure of the genetic material of the human body and various pathogens, so as to achieve the purpose of predicting and diagnosing diseases.
In recent years, with the upgrading and iteration of molecular diagnostic technology, the clinical application of molecular diagnostics has become more and more extensive and in-depth, and the molecular diagnostics market has entered a period of rapid development.
The author summarizes the common molecular diagnostic technologies on the market, and is divided into three parts: the first part introduces the PCR technology, the second part introduces the nucleic acid isothermal amplification technology, and the second part introduces the sequencing technology.
01
Part I: PCR Technology
PCR technology
PCR (polymerase chain reaction) is one of the in vitro DNA amplification technologies, with a history of more than 30 years.
PCR technology was pioneered in 1983 by Kary Mullis of Cetus, USA. Mullis applied for a PCR patent in 1985 and published the first PCR academic paper on Science in the same year. Mullis won the Nobel Prize in Chemistry in 1993.
Basic Principles of PCR
PCR can amplify target DNA fragments by more than one million times. The principle is that under the catalysis of DNA polymerase, the parent strand DNA is used as a template, and a specific primer is used as the starting point for extension. It is replicated in vitro through steps such as denaturation, annealing, and extension. The process of daughter strand DNA complementary to the parent strand template DNA.
The standard PCR process is divided into three steps:
1. Denaturation: Use high temperature to separate DNA double strands. The hydrogen bonds between DNA double strands are broken at high temperatures (93-98°C).
2. Annealing: After the double-stranded DNA is separated, the temperature is lowered so that the primer can bind to the single-stranded DNA.
3. Extension: The DNA polymerase starts to synthesize complementary strands along the DNA strands from the primers bound when the temperature is lowered. When the extension is completed, a cycle is completed, and the number of DNA fragments doubles.
Reciprocating these three steps 25-35 times, the number of DNA fragments will increase exponentially.
The ingenuity of PCR is that different primers can be designed for different target genes, so that the target gene fragments can be amplified in a short period of time.
So far, PCR can be divided into three categories, namely ordinary PCR, fluorescent quantitative PCR and digital PCR.
The first generation of ordinary PCR
Use an ordinary PCR amplification instrument to amplify the target gene, and then use agarose gel electrophoresis to detect the product, only qualitative analysis can be done.
The main disadvantages of the first generation PCR:
-Prone to non-specific amplification and false positive results.
-The detection takes a long time and the operation is cumbersome.
-Only qualitative testing can be done.
Second-generation fluorescence quantitative PCR
Fluorescence quantitative PCR (Real-Time PCR), also known as qPCR, is used to monitor the accumulation of amplified products through the accumulation of fluorescent signals by adding fluorescent probes that can indicate the progress of the reaction system, and to judge the results through the fluorescence curve, and It can be quantified with the help of Cq value and standard curve.
Because the qPCR technology is carried out in a closed system, the probability of contamination is reduced, and the fluorescence signal can be monitored for quantitative detection, so it is the most widely used in clinical practice and has become the dominant technology in PCR.
The fluorescent substances used in real-time fluorescent quantitative PCR can be divided into: TaqMan fluorescent probes, molecular beacons and fluorescent dyes.
1) TaqMan fluorescent probe:
During PCR amplification, a specific fluorescent probe is added while adding a pair of primers. The probe is an oligonucleotide, and the two ends are respectively labeled with a reporter fluorescent group and a quencher fluorescent group.
When the probe is intact, the fluorescent signal emitted by the reporter group is absorbed by the quenching group; during PCR amplification, the 5′-3′ exonuclease activity of Taq enzyme cleaves and degrades the probe, making the reporter fluorescent group and quencher The fluorescent group is separated, so that the fluorescence monitoring system can receive the fluorescence signal, that is, every time a DNA strand is amplified, a fluorescent molecule is formed, and the accumulation of the fluorescence signal is completely synchronized with the formation of the PCR product.
2) SYBR fluorescent dyes:
In the PCR reaction system, an excess of SYBR fluorescent dye is added. After the SYBR fluorescent dye is non-specifically incorporated into the DNA double-strand, it emits a fluorescent signal. The SYBR dye molecule that is not incorporated into the chain will not emit any fluorescent signal, thereby ensuring the fluorescent signal The increase in PCR products is completely synchronized with the increase in PCR products. SYBR only binds to double-stranded DNA, so the melting curve can be used to determine whether the PCR reaction is specific.
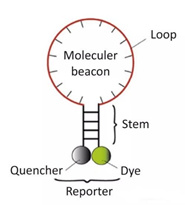
3) Molecular beacons
It is a stem-loop double-labeled oligonucleotide probe that forms a hairpin structure of about 8 bases at the 5 and 3 ends. The nucleic acid sequences at both ends are complementarily paired, causing the fluorescent group and the quenching group to be tight. Close, it will not produce fluorescence.
After the PCR product is generated, during the annealing process, the middle part of the molecular beacon is paired with a specific DNA sequence, and the fluorescent gene is separated from the quencher gene to produce fluorescence.
The main disadvantages of second-generation PCR:
Sensitivity is still lacking, and the detection of low-copy specimens is not accurate.
There is background value influence, and the result is susceptible to interference.
Third generation digital PCR
Digital PCR (DigitalPCR, dPCR, Dig-PCR) calculates the copy number of the target sequence through end-point detection, and can perform accurate absolute quantitative detection without using internal controls and standard curves.
Digital PCR uses endpoint detection and does not depend on the Ct value (cycle threshold), so the digital PCR reaction is less affected by the amplification efficiency, and the tolerance to PCR reaction inhibitors is improved, with high accuracy and reproducibility.
Due to the characteristics of high sensitivity and high accuracy, it is not easily interfered by PCR reaction inhibitors, and it can achieve true absolute quantification without standard products, which has become a research and application hotspot.
According to the different forms of the reaction unit, it can be divided into three types: microfluidic, chip and droplet systems.
Post time: Jul-08-2021