1. Detect the absorbance of RNA solution
Absorbance at 280, 320, 230, and 260 nm represents the values of nucleic acid, background (solution turbidity), salt concentration, and organic matter such as protein, respectively. Generally only look at OD260/OD280 (Ratio, R). When 1.8~2.0, we think that the contamination of protein or other organic matter in RNA can be tolerated, but it should be noted that when Tris is used as the buffer to detect the absorbance, the R value may be greater than 2 (generally it should be <2.2). When R<1.8, the pollution of protein or other organic matter in the solution is more obvious, and the fate of the RNA can be determined according to the needs. When R>2.2, it means that RNA has been hydrolyzed into single nucleic acid.
2.Electrophoretic pattern of RNA
Generally, denaturing gel is used for RNA electrophoresis, but if it is only for detecting the quality of RNA, denaturing gel is not necessary, and ordinary agarose gel can be used. The purpose of electrophoresis is to detect the integrity of 28S and 18S bands and their ratio, or the integrity of mRNA smear. Generally, if the 28S and 18S bands are bright, clear, and sharp (referring to the edges of the bands are clear), and the brightness of 28S is more than twice that of the 18S band, we consider the quality of the RNA to be good.
The above are the two methods we commonly use, but neither of these two methods can clearly tell us whether there is residual RNase in the RNA solution. If there is a very small amount of RNase in the solution, it is difficult for us to detect it with the above method, but most of the subsequent enzymatic reactions are carried out at above 37 degrees and for a long time. In this way, if there is a very small amount of RNase in the RNA solution, then there will be a very suitable environment and time to play their role in the subsequent experiments, and of course the experiment will be cold at this time. Below we introduce a method that can confirm whether there is residual RNase in the RNA solution.
3. Heat preservation test
According to the sample concentration, draw two 1000 ng RNA from the RNA solution and add it to a 0.5 ml centrifuge tube, and supplement it with pH 7.0 Tris buffer to a total volume of 10 ul, and then seal the cap of the tube. Put one of them in a constant temperature water bath at 70°C and keep it warm for 1 h. The other part was stored in a -20°C refrigerator for 1 h. When the time is up, remove the two samples for electrophoresis. After the electrophoresis is completed, compare the electrophoretic bands of the two. If the bands of the two are consistent or have no significant difference (of course, their bands also meet the conditions in method 2), it means that there is no residual RNase contamination in the RNA solution, and the quality of the RNA is very good. On the contrary, if the sample incubated at 70°C shows obvious degradation, it indicates that there is RNase contamination in the RNA solution.
2 Experimental methods and techniques for RNA extraction
The problems we often encounter when extracting RNA are: (1) RNA yield is low; (2) RNA has serious salt pollution; (3) RNA has serious organic solvent pollution; (4) sample degradation and other problems
1. Commonly used total RNA extraction reagents
The guanidine isothiocyanate method and the Trizol method are the most commonly used methods for the extraction of total RNA from animal tissues and animal cells. It is especially suitable for small samples and tissues that are particularly difficult to extract, such as the extraction of total RNA from rabbit skin and animal connective tissue; in addition, Trizol, as a general-purpose lysis reagent, can also be used for the extraction of plant tissues, bacteria, fungi and other tissues. For plant tissues containing polysaccharides and polyphenols, such as camellia oleifera, tea leaves, rapeseed, etc., the CTAB method can also be used to extract total RNA.
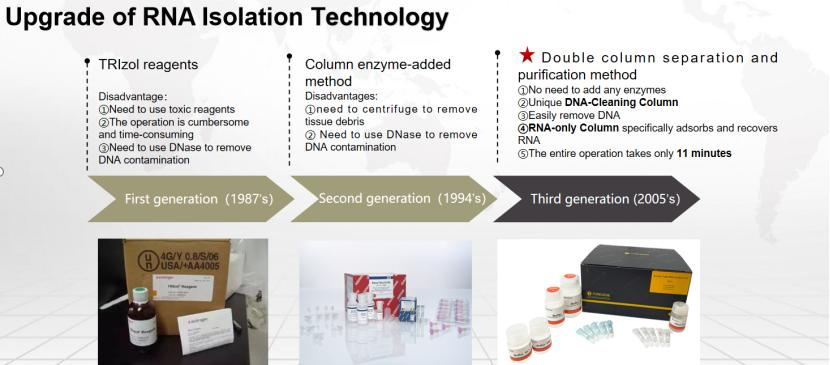
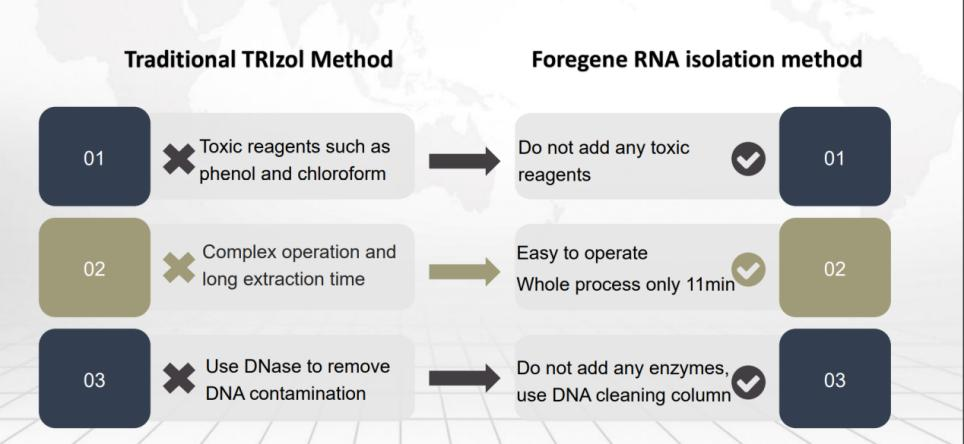
As a conventional method, the double-column method is also very popular due to its normal temperature operation, no need to add RNase, and safety—no chloroform, phenols and other organic reagents for extraction. (recommended products )
2. Extraction of total RNA from animal tissues
(1) Try to choose fresh tissue, if it is not fresh (preferably within three months - 80 ℃ refrigerator or frozen in liquid nitrogen. When cutting tissue, do not cut directly at room temperature, be sure to Put it on the ice box, try to avoid repeated freezing and thawing.
(2) Use clean scissors and tweezers to cut a small piece of tissue, try to cut the central part of the tissue when cutting the sample, or first cut the large piece of tissue from the middle, and then cut the sample at the fresh incision position. The removed tissue should be fully shredded, put the shredded tissue into an EP tube without RNase, add the lysate, the shredded tissue should be fully exposed to the lysate, and prepare for homogenization.
(3) For normal tissues, select mung bean-sized tissues (30-60 mg) for homogenization. If the tissues contain a large amount of protein, fat, or dense fibrous tissues such as liver, appropriately increase or decrease the amount of cut tissues (optional) Choose 10~20 mg).
(4) If fish muscle, shrimp meat, jellyfish and other tissues with high water content are extracted, the sample volume should be appropriately increased (recommended 100-200 mg).
(5) If conditions permit, the animal tissue can be directly extracted after being homogenized with a high-passage tissue homogenizer, if there is no such equipment.
(6) The RNA obtained after the final extraction must be placed on the ice box immediately to reduce the degradation of RNA.
3. Animal cell RNA extraction
(1) Suspension cells: centrifuge directly and discard the medium, wash with sterile PBS for 1-2 times, then suspend with an appropriate amount of PBS, and then add lysate for lysis. Do not add the lysate directly to the precipitated cells after completely discarding the liquid. This will cause the histone package released after the lysed cells on the outer layer to adhere to the outside of the precipitated cells, thereby limiting the contact of the cells inside the pellet with the lysate. , resulting in incomplete cell lysis and reduced RNA yield.
(2) Cells that are semi-adherent or not tightly adherent: After discarding the medium, wash with PBS for 1-2 times, then directly absorb an appropriate amount of PBS and blow the culture dish with a pipette or gun to blow off the cells, and transfer them to RNA-free cells. Add the lysate to the EP tube of the enzyme for extraction.
(3) Adherent cells: need to be digested with trypsin first, then collected into RNase-free EP tubes, centrifuged to remove the supernatant, washed 1-2 times with PBS to remove excess trypsin, and resuspended with an appropriate amount of PBS Then proceed to the extraction step.
4. Plant RNA extraction
Plant tissues are rich in phenolic compounds, or rich in polysaccharides, or contain some unidentified secondary metabolites, or have high activity of RNase. These substances are tightly combined with RNA after cell lysis to form insoluble complexes or colloidal precipitates, which are difficult to remove. Therefore, when we extract plant tissue, we need to choose a kit for plants. The lysate in the kit can effectively solve the problems of easy oxidation of polyphenols and separation of polysaccharide compounds and nucleic acids.
(For polysaccharide polyphenol plant RNA extraction, recommended products:
(1) The peel, pulp, seeds, leaves, etc. of the plant should be fully ground in a mortar. During the grinding process, liquid nitrogen should be replenished in time to avoid melting the sample. The ground sample should be quickly added to the lysate and shaken to avoid RNA degradation.
(2) For fiber-rich samples such as rice and wheat leaves, the amount of extraction should be appropriately reduced, otherwise the tissue grinding and lysis will not be complete, resulting in a low yield of extracted RNA.
(3) For plant tissues with high water content, such as pomegranate fruit, watermelon fruit, peach fruit, etc., the sample size should be appropriately increased (100-200 mg is optional).
(4) Plant tissues, such as plant leaves, rhizomes, hard fruits and other materials are generally recommended to use liquid nitrogen to thoroughly mortar the ingredients in a mortar, and then proceed to the extraction step. Conventional tissue homogenizers may not be effective in homogenizing plant tissues, and are generally not recommended.
5. Precautions for RNA extraction
(1) Tissue samples should be as fresh as possible to avoid repeated freezing and thawing.
(2) The tissue should be fully ground during extraction, and the amount of tissue should not be too little, let alone too much.
(3) Sufficient incubation time should be given after adding the lysate to fully lyse the sample.
(4) When using the Trizol method for extraction, the principle of absorbing the supernatant after stratification is "prefer to inhale less than inhale more", and must not extract to the middle layer, otherwise it will cause serious genomic DNA contamination.
(5) When washing, the washing liquid should fully infiltrate around the tube wall to ensure thorough washing.
(6) For the column extraction method, in addition to detaching the column after washing, the adsorption column should also be placed in an ultra-clean bench and blown for 5-10 minutes to fully evaporate the organic solvent to dryness.
(7) At the last elution of the column method, after adding DEPC water, it should be incubated for 3-5 minutes, or the DEPC water should be heated to 60°C in advance to increase the elution yield. In the traditional Trizol cleavage and isopropanol precipitation method, the final RNA is dissolved in DEPC water, so an appropriate time should be given for dissolution, and the bottom of the centrifuge tube should be continuously blown with a pipette tip.
3 Three Causes and solutions for low RNA concentration/poor quality
1. The yield is too low
The extracted sample is too low, the total amount is insufficient, or the extracted sample is too much and the lysis is not complete; the tissue or cells of appropriate quality should be used for extraction, the pre-treatment of the sample must be done well, and the lysis should be sufficient.
2. Genome residues
When extracting by Trizol method, when the supernatant is sucked into the middle layer after layering, serious genome contamination will be caused; extra care should be taken when layering to avoid sucking into the middle layer. If the column method is used for extraction, a kit containing DNase I can be selected for extraction. The nucleic acid adsorbed on the membrane is directly digested with DNase I, which can greatly reduce DNA residues.
3. RNA degradation
It may be the degradation of the extracted sample itself, or the degradation caused during the extraction process; as far as possible, fresh samples should be used for RNA extraction, and the collected samples should be stored in liquid nitrogen or -80°C refrigerator in time, and repeated freezing and thawing should be avoided. RNase/DNase free tips, centrifuge tubes and other materials should be used in the RNA extraction process. The extraction process should be as fast as possible. The extracted RNA should be placed on an ice box and stored at -80 in time. If the extracted RNA needs to be detected by gel electrophoresis, electrophoresis should be performed immediately after extraction, and the electrophoresis buffer should be replaced with a newly prepared one.
4. Salt and organic solvent residues
The extraction reagents contain phenol and guanidine salts, and the washing solution contains ethanol. During the extraction process, the lysate was not completely absorbed and discarded, and the washing solution was not fully dried. Residual salts and organic solvents are harmful to subsequent reverse transcription and PCR. Different degrees of inhibition, so the tissue lysate should be fully removed during the extraction process, and the washing should be sufficient so that the surrounding walls of the tube can be washed. In addition, the tube is emptied and blown is a necessary step, which will further reduce the residue of organic matter.
For more information about RNA extraction, please follow our website:
www.foreivd.com for more information.

Post time: Dec-01-2022