In the past ten years, the gene editing technology based on CRISPR has developed rapidly, and has been successfully applied to the treatment of genetic diseases and cancer in human clinical trials. At the same time, scientists around the world are constantly tap new new tools with gene editing potential to solve the problems of existing gene editing tools and decisives.
In September 2021, Zhang Feng’s team published a paper in the Science journal [1], and found that a wide range of transposters coded RNA guided nucleic acid enzymes and named it Omega system (including ISCB, ISRB, TNP8). The study also found that the Omega system uses a section of RNA to guide the cutting DNA dual chain, namely ωRNA. More importantly, these nucleic acid enzymes are very small, only about 30%of CAS9, which means that they may be more likely to be delivered to cells.

On October 12, 2022, Zhang Feng’s team published in the Nature journal entitled: Structure of the Omega Nickase ISRB in Complex with ωrna and Target DNA [2].
The study further analyzed the frozen electron microscope structure of ISRB-ωRNA and target DNA complex in the Omega system.
ISCB is the ancestor of CAS9, and ISRB is the same object of the lack of the HNH nucleic acid domain of ISCB, so the size is smaller, only about 350 amino acids. DNA also provides the foundation for further development and engineering transformation.

RNA-guided IsrB is a member of the OMEGA family encoded by the IS200/IS605 superfamily of transposons. From phylogenetic analysis and shared unique domains, IsrB is likely to be the precursor of IscB, which is the ancestor of Cas9.
In May 2022, Cornell University’s Lovely Dragon Laboratory published a paper in the journal Science [3], analyzing the structure of IscB-ωRNA and its mechanism of cutting DNA.

Compared with IscB and Cas9, IsrB lacks the HNH nuclease domain, the REC lobe, and most of the PAM sequence-interacting domains, so IsrB is much smaller than Cas9 (only about 350 amino acids). However, the small size of IsrB is balanced by a relatively large guide RNA (its omega RNA is about 300 nt long).
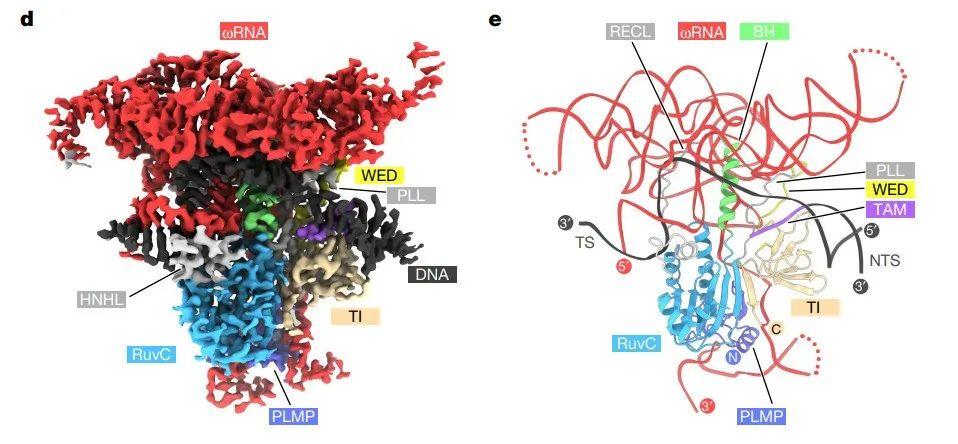
Zhang Feng’s team analyzed the cryo-electron microscope structure of IsrB (DtIsrB) from the damp-heat anaerobic bacterium Desulfovirgula thermocuniculi and its complex of ωRNA and target DNA. Structural analysis showed that the overall structure of IsrB protein shared a backbone structure with Cas9 protein.
But the difference is that Cas9 uses the REC lobe to facilitate target recognition, while IsrB relies on its ωRNA, a part of which forms a complex three-dimensional structure that acts like REC.
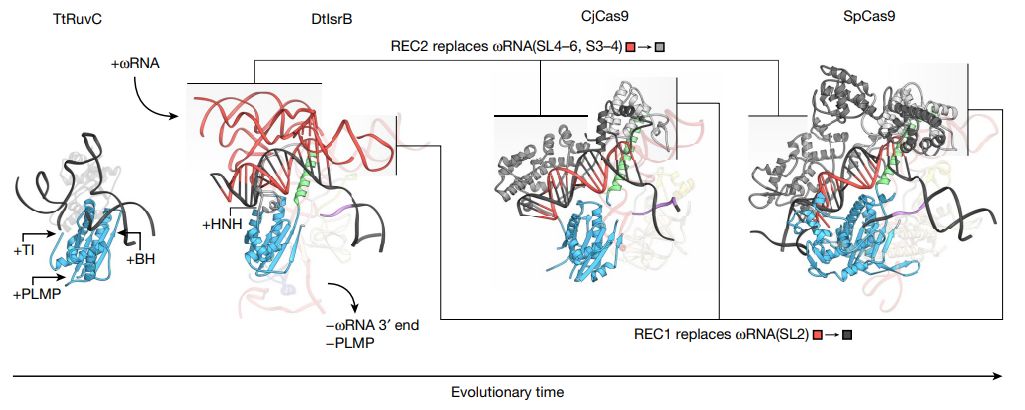
To better understand the structural changes of IsrB and Cas9 during the evolution from RuvC, Zhang Feng’s team compared the target DNA-binding structures of RuvC (TtRuvC), IsrB, CjCas9 and SpCas9 from Thermus thermophilus .
The structural analysis of IsrB and its ωRNA clarifies how IsrB-ωRNA jointly recognizes and cleaves target DNA, and also provides a basis for further development and engineering of this miniaturized nuclease. Comparisons with other RNA-guided systems highlight functional interactions between proteins and RNAs, advancing our understanding of the biology and evolution of these diverse systems.
Links:
1.https://www.science.org/doi/10.1126/science.abj6856
2.https://www.science.org/doi/10.1126/science.abq7220
3.https://www.nature.com/articles/s41586-022-05324-6
Post time: Oct-14-2022








