Several revolutionary inventions in the history of detection technology in my mind are immunolabeling technology based on the principle of antigen-antibody specific binding, PCR technology and sequencing technology. Today we will talk about PCR technology. According to the evolution of PCR technology, people habitually divide PCR technology into three generations: ordinary PCR technology, real-time fluorescent quantitative PCR technology and digital PCR technology.
Common PCR technique

KARY MULLIS (1944.12.28-2019.8.7)
Kary Mullis invented the polymerase chain reaction (polymerase chain reaction , PCR) in 1983. It is said that when he was driving his girlfriend, he suddenly had a flash of inspiration and thought of the principle of PCR (on the benefits of driving). Kary Mullis was awarded the Nobel Prize in Chemistry in 1993. The New York Times commented: "Highly original and significant, almost dividing biology into pre-PCR and post-PCR eras.
The principle of PCR: Under the catalysis of DNA polymerase, the mother strand DNA is used as the template, and the specific primer is used as the starting point of extension, and the daughter strand DNA complementary to the mother strand template DNA is copied in vitro through denaturation, annealing, extension and other steps. It is a DNA synthesis amplification technology in vitro, which can rapidly and specifically amplify any target DNA in vitro.
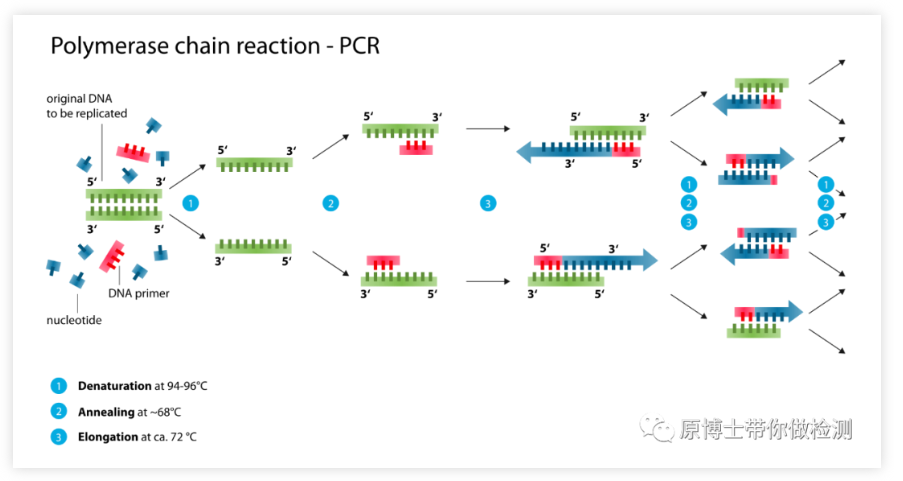
Advantages of ordinary PCR
1.Classic method, complete international and domestic standards
2.Lower cost of instrument reagents
3.PCR products can be recovered for other molecular biology experiments
Recommended Foregene PCR machine: https://www.foreivd.com/foreamp-sn-695-series-thermal-cycler-96-wells-pcr-machine-product/
Related products: https://www.foreivd.com/pcr-herotm-with-dye-product/
Disadvantages of ordinary PCR
1.easy to pollute
2.cumbersome operation
3.only qualitative analysis
4.Moderate sensitivity
5.There is non-specific amplification, and when the non-specific band is the same size as the target band, it cannot be distinguished
Capillary electrophoresis-based PCR
In response to the shortcomings of ordinary PCR, some manufacturers have introduced instruments based on the principle of capillary electrophoresis. The electrophoresis step after PCR amplification is completed in the capillary. The sensitivity is higher, and the difference of several bases can be distinguished and the amplification can be calculated by MAERKER. product content. The disadvantage is that the PCR product still needs to be opened and put into the instrument, and there is still a great risk of contamination.

Capillary Electrophoresis
2. Real-time fluorescent quantitative PCR (Quantitative Real-time PCR, qPCR) technology Fluorescent quantitative PCR, also called Real-Time PCR, is a new nucleic acid quantitative technology developed by PE (Perkin Elmer) in 1995. The development history of fluorescent quantitative PCR is a history of soul-stirring struggles of giants such as ABI, Roche, and Bio-Rad. If you are interested, you can check it out. This technique is currently the most mature and widely used semi-quantitative PCR technique.
Recommended qPCR Machine:https://www.foreivd.com/mini-real-time-pcr-system-forequant-sf2sf4-product/
Fluorescent dye method (SYBR Green I): SYBR Green I is the most commonly used DNA-binding dye for quantitative PCR, which binds non-specifically to double-stranded DNA. In the free state, SYBR Green emits weak fluorescence, but once bound to double-stranded DNA, its fluorescence increases 1000-fold. Therefore, the total fluorescent signal emitted by a reaction is proportional to the amount of double-stranded DNA present and will increase with the increase of amplified product. Since the dye binds non-specifically to double-stranded DNA, false positive results may be generated.
Related products: https://www.foreivd.com/real-time-pcr-easytm-sybr-green-i-kit-product/
Fluorescent probe method (Taqman technology): During PCR amplification, a specific fluorescent probe is added at the same time as a pair of primers. The probe is a linear oligonucleotide, with a fluorescent reporter group and a fluorescent quencher group respectively labeled at both ends. When the probe is intact, the fluorescent signal emitted by the reporter group is absorbed by the quencher group, and the detection There is no fluorescent signal; during PCR amplification (in the extension stage), the 5'-3' Dicer activity of Taq enzyme will digest and degrade the probe, so that the reporter fluorescent group and the quencher fluorescent group are separated, so that the fluorescence monitoring system The fluorescent signal can be received, that is, every time a DNA chain is amplified, a fluorescent molecule is formed, which realizes the complete synchronization of the accumulation of fluorescent signals and the formation of PCR products. Taqman probe method is the most commonly used detection method in clinical detection.
Related products: https://www.foreivd.com/quickeasy%e1%b5%80%e1%b4%b9-real-time-pcr-kit-taqman-product/
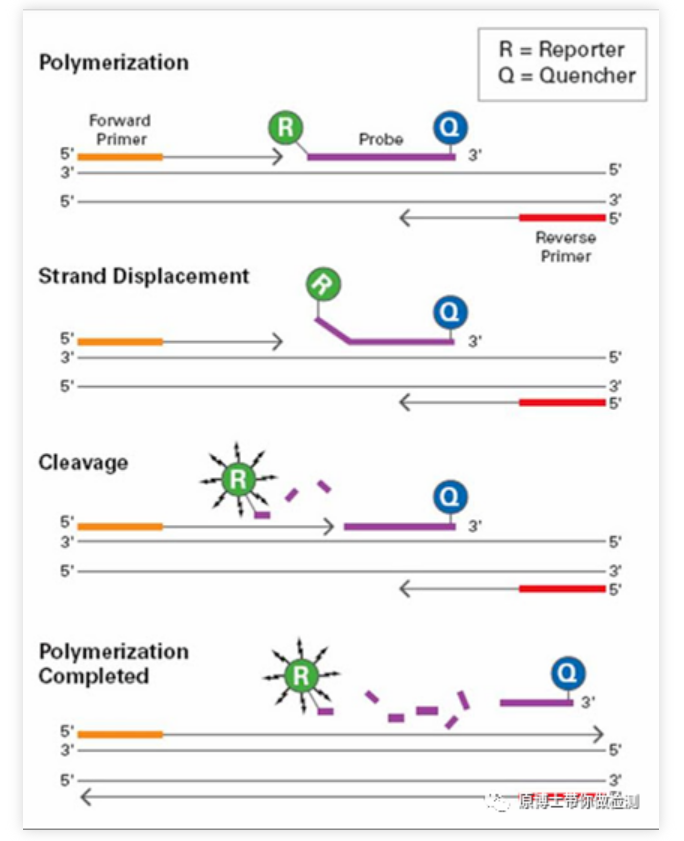
Advantages of qPCR
1.The method is mature and the supporting equipment and reagents are complete
2.Medium cost of reagents
3.easy to use
4.High detection sensitivity and specificity
Disadvantages of qPCR
The mutation of the target gene leads to missed detection.
The detection result of low-concentration template cannot be determined.
There is a large error when using the standard curve for quantitative detection.
3. Digital PCR (digital PCR, dPCR) technology
Digital PCR is a technique for the absolute quantification of nucleic acid molecules. Compared with qPCR, digital PCR can directly read the number of DNA/RNA molecules, which is the absolute quantification of nucleic acid molecules in the starting sample. In 1999, Bert Vogelstein and Kenneth W. Kin-zler formally proposed the concept of dPCR.
In 2006, Fluidigm was the first to produce a commercial chip-based dPCR instrument. In 2009, Life Technologies launched the OpenArray and QuantStudio 12K Flex dPCR systems. In 2013, Life Technologies launched the QuantStudio 3DdPCR system, which uses high-density nanoscale microfluidic chip technology to evenly distribute samples to 20,000 individual cells. in the reaction well.
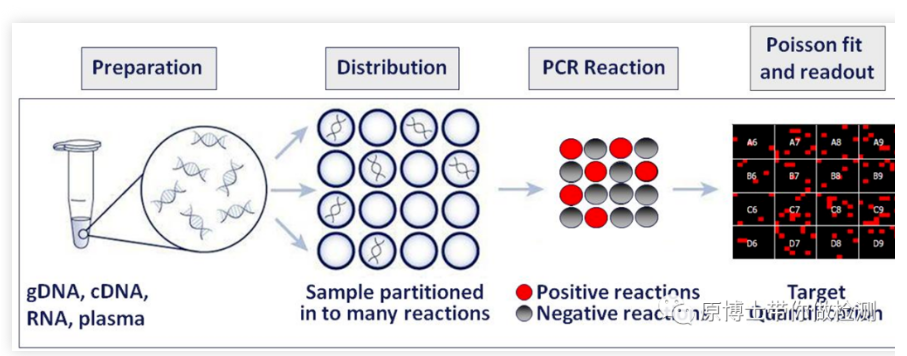
In 2011, Bio-Rad launched the droplet-based QX100 dPCR instrument, which uses water-in-oil technology to evenly distribute the sample to 20,000 droplet-water-in-oil, and uses a droplet analyzer to analyze the droplets. In 2012, RainDance launched the RainDrop dPCR instrument, driven by high-pressure gas, to divide each standard reaction system into a reaction emulsion containing 1 million to 10 million picoliter-level micro-droplets.
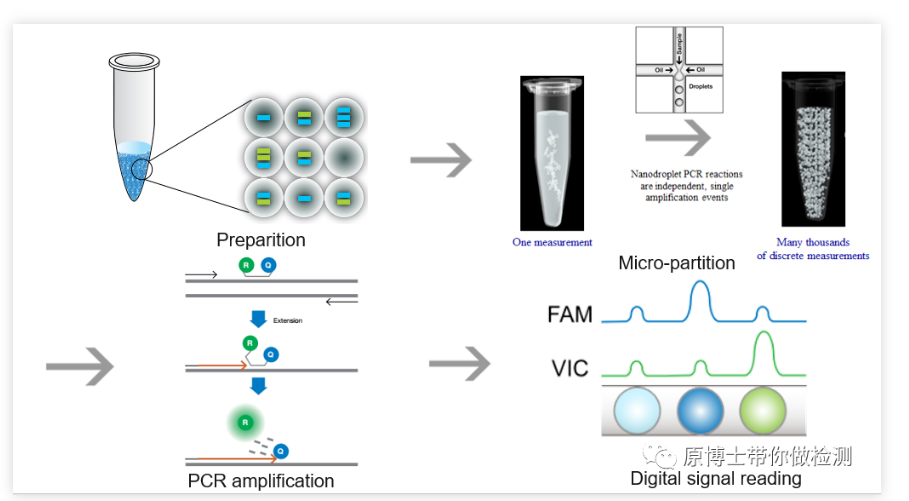
So far, digital PCR has formed two major factions, chip type and droplet type. No matter what kind of digital PCR, its core principles are limiting dilution, endpoint PCR and Poisson distribution. The standard PCR reaction system containing nucleic acid templates is evenly divided into tens of thousands of PCR reactions, which are distributed to chips or microdroplets, so that each reaction contains as much as possible a template molecule, and a single-molecule template PCR reaction is performed. By reading the fluorescence The presence or absence of the signal is counted, and the absolute quantification is performed after calibration of the statistical Poisson distribution.
The following are the characteristics of several digital PCR platforms I have used:
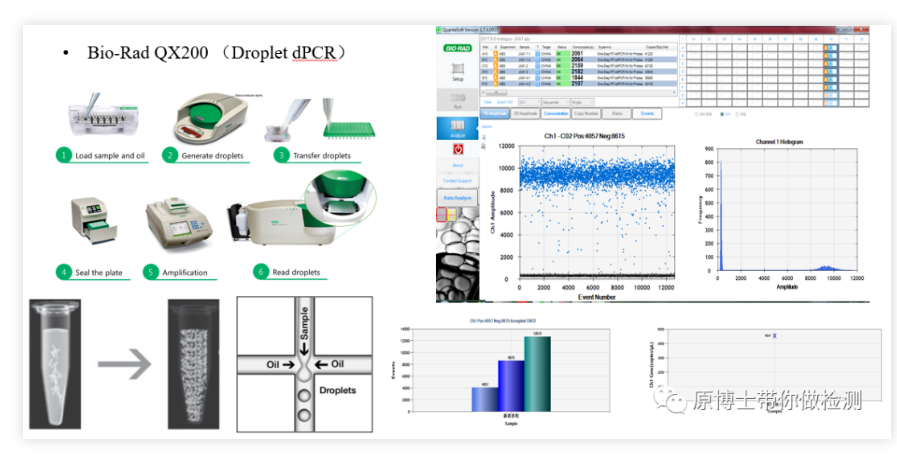
1. Bio-Rad QX200 droplet digital PCR Bio-Rad QX200 is a very classic digital PCR platform, the basic detection process: 20,000 samples are generated by the droplet generator Water-in-oil micro-droplets are amplified on an ordinary PCR machine, and finally the fluorescence signal of each micro-droplet is read by a micro-droplet reader. The operation is more complicated, and the risk of pollution is medium.
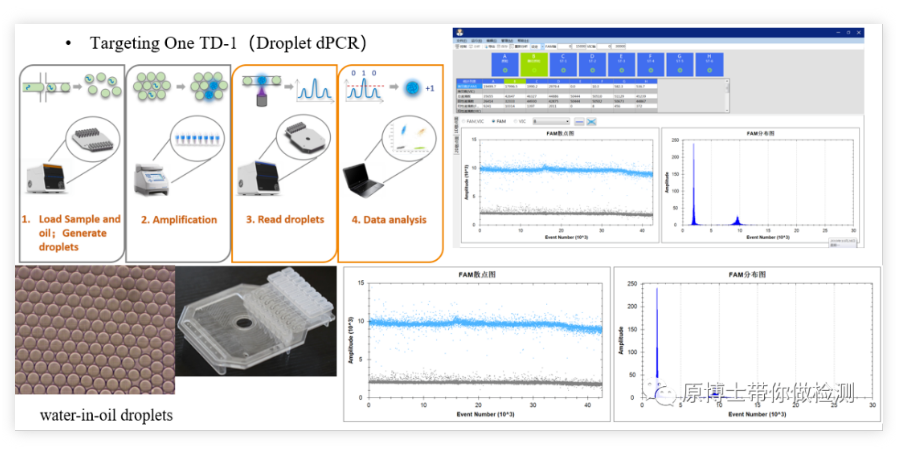
Xinyi TD1 micro-droplet digital PCR Xinyi TD1 is a domestic digital PCR platform, the basic detection process: generate 30,000-50,000 water-in-oil droplets through a droplet generator, amplify on a common PCR instrument, and finally pass The droplet reader reads the fluorescent signal of each droplet. Both droplet generation and reading in this platform are performed in a dedicated chip with low risk of contamination.
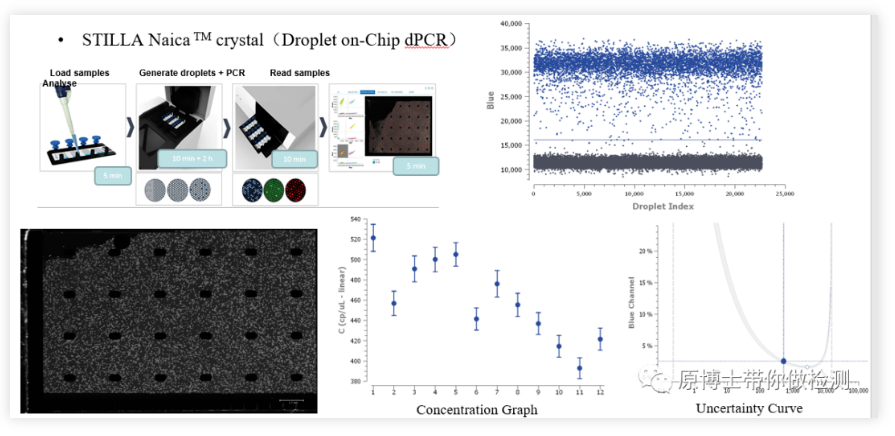
STILLA Naica micro-droplet chip digital PCR STILLA Naica is a relatively new digital PCR platform. The basic detection process is: add the reaction solution to the chip, put the chip into the micro-droplet generation and amplification system, and generate 30,000 micro-droplets. Spread on the chip, and PCR amplification is completed on the chip. Then the amplified chip is transferred to the micro-droplet reading analysis system, and the fluorescent signal is read by taking pictures. Since the entire process takes place in a closed chip, the risk of contamination is low.
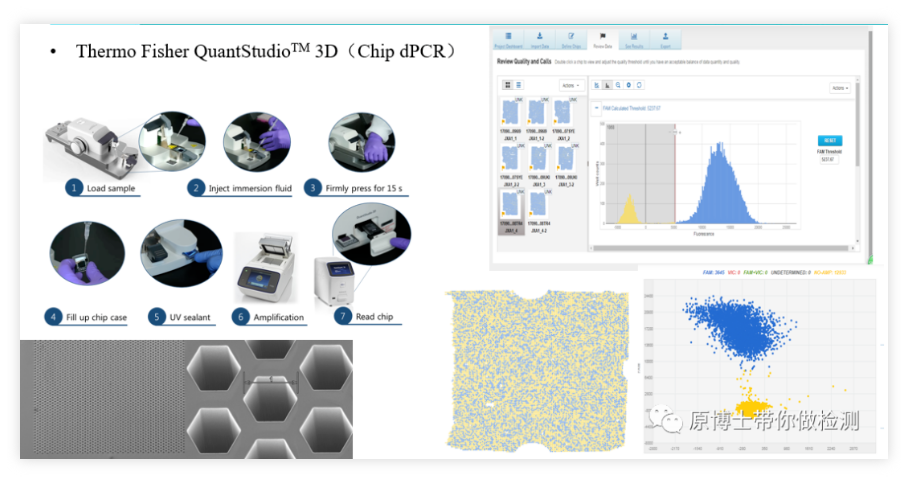
4. ThermoFisher QuantStudio 3D chip digital PCR
ThermoFisher QuantStudio 3D is another classic chip-based digital PCR platform. Its basic detection process is: add the reaction solution into the spreader, and spread the reaction solution evenly on the chip with 20,000 microwells through the spreader. , put the chip on the PCR machine to amplify, and finally put the chip into the reader and take a photo to read the fluorescent signal. The operation is relatively complicated, and the whole process is carried out in a closed chip, and the risk of contamination is low.
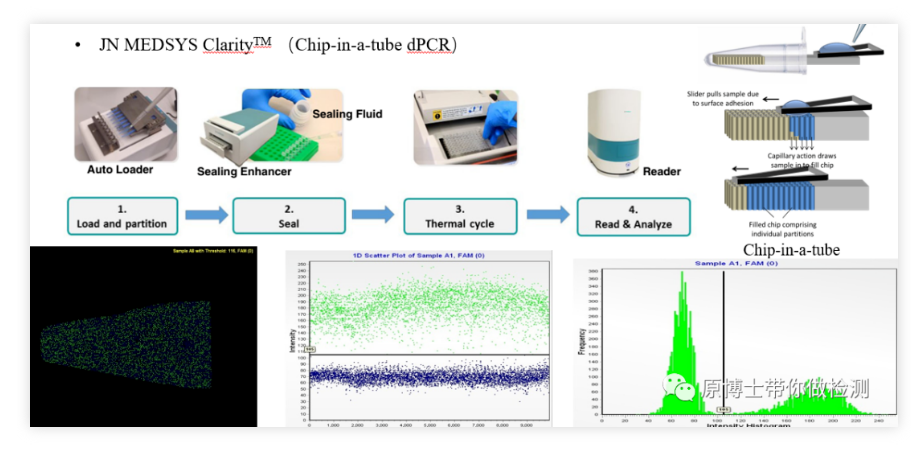
5. JN MEDSYS Clarity chip digital PCR
JN MEDSYS Clarity is a relatively new chip-type digital PCR platform. Its basic detection process is: add the reaction solution into the applicator, and spread the reaction solution evenly on 10,000 PCR tubes fixed in the PCR tube through the applicator. On the microporous chip, the reaction solution enters the chip through capillary action, and the PCR tube with the chip is placed on the PCR machine for amplification, and finally the chip is put into the reader to read the fluorescent signal by taking a photo. The operation is more complicated. The risk of contamination is low.
The parameters of each digital PCR platform are summarized as follows:
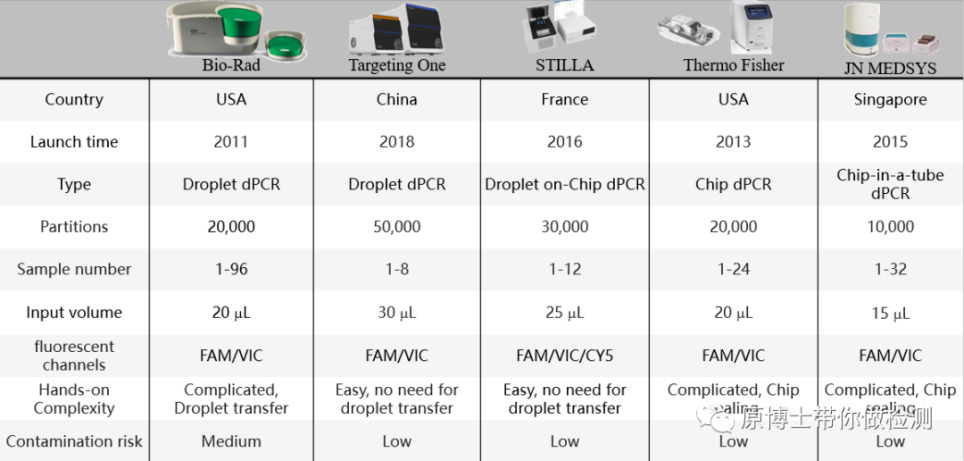
The evaluation indicators of the digital PCR platform are: the number of split units, the number of fluorescent channels, the complexity of operation and the risk of contamination. But the most important thing is the detection accuracy. One way to evaluate digital PCR platforms is to use multiple digital PCR platforms to verify each other, and another way is to use standard substances with accurate values.
Advantages of dPCR
1.Achieving absolute quantification
2.Higher sensitivity and specificity
3.Can detect low copy samples
Disadvantages of dPCR 1. Expensive equipment and reagents 2. Complicated operation and long detection time 3. Narrow detection range
At present, the three generations of PCR technology have their own advantages and disadvantages, and each has its own application fields, and it is not a relationship that one generation replaces the other. The continuous advancement of technology has injected new vitality into PCR technology, enabling it to unlock one application direction after another, making nucleic acid detection more convenient and accurate.
Source: Dr. Yuan takes you for testing
Recommended Products:
Post time: Nov-18-2022











