Everyone is talking about the principle of qRT-PCR experiment, primer design, result interpretation, etc., but I think I should share with you the experimental operation of qRT-PCR. It’s small, but it’s about results.
Before doing qRT-PCR, we need to have a clear understanding of our own RNA and operation methods. After all, our efforts are aimed at getting results, rather than simply practicing. So before doing qRT-PCR, we need to determine the following issues (some of which are only applicable to SYBR).
1 Are you sure your RNA is not degraded?
NanoDrop 2000 can only detect the concentration and purity of RNA, but cannot detect the integrity of RNA.
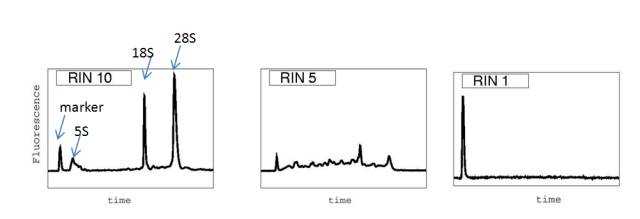
The RNA (RNA Intesity Number) value can reflect the integrity of the RNA, which is detected by the Agilent 2100 Bioanalyzer system.
Fig. Schematic diagram of RIN values for different RNA samples (eukaryotes)
However, laboratories generally do not have Agilent 2100 Bioanalyzer. In this case, we can detect through formaldehyde gel, but the requirement for the total amount of RNA is high, so the fastest method is to use ordinary gel electrophoresis. It is required to be in a nuclease-free environment, so it is necessary to rinse the electrophoresis tank, sol bottle, gel bracket and comb with DEPC water. The agarose is also nuclease-free (as long as it is freshly opened), and the Loading Buffer should be freshly opened as much as possible, with 1.2% gel.
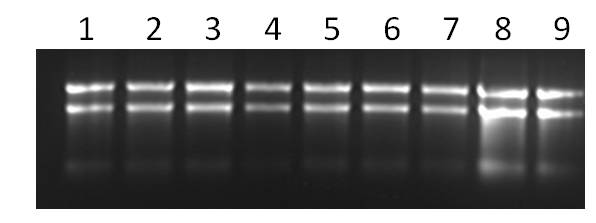
Note that the gel must be completely dissolved, otherwise it will cause inhomogeneous bands, as shown in sample 9 in the figure. If the voltage is too high or running for too long will generate heat and cause RNA degradation, so the voltage and time should be controlled reasonably. In addition, gel running can also further determine whether there is DNA residue in the sample, and observe whether there are a large number of retained bands in the dispensing well.
Figure. Gel electrophoresis detection of RNA
2 Are you sure about the concentration of your cDNA?
The experience of the big brothers in the laboratory is that the cDNA of the 20 ul system obtained by each inversion is directly diluted 20X, while the post-doctoral sisters are diluted 10X. I usually depend on the situation. Because the quality of the RNA mentioned by each person is different, the level of reversal is also different, and the reversal technology may not be stable.
So every time I get the reversed cDNA, I will first dilute it about 3 times, and then use the housekeeping gene to do RT-PCR, the number of cycles is generally 25 cycles, to identify the specific concentration, and then determine the final dilution factor.
3 Are you sure your primers are easy to use?
It can pass the melting curve of qRT-PCR, but this still costs money. For laboratories without a lot of money, when they get a lot of primers, they can use ordinary RT-PCR to see if it is a single band and identify the specificity of the primers. If the laboratory is not short of money, the specificity of all primers can be identified once through the melting curve.
4 Are you sure that your experimental conditions are suitable?
SYBR should be protected from strong light, so try to turn off the overhead light when adding SYBR reagent, and only need to use dim light to complete it.
Store SYBR at 4°C. When in use, gently invert up and down to mix well to avoid foaming, and do not vortex vigorously.
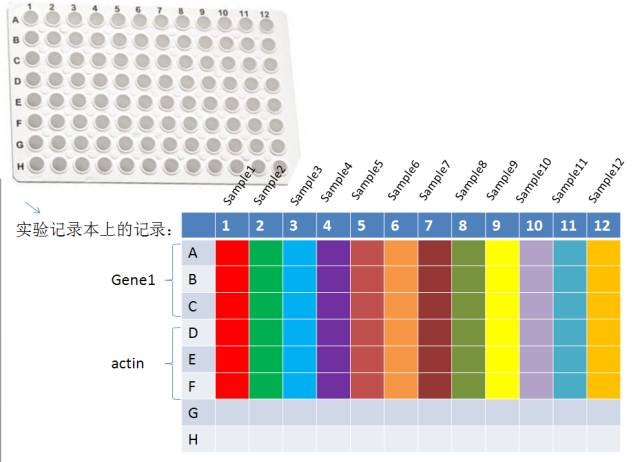
Some younger sisters like to draw marks on the PCR board for fear of mixing up the samples, which is wrong. Because your markers are very likely to affect the collection of fluorescent signals, I generally recommend juniors to use experimental notebooks to assist in memory, as shown below.
Figure. qRT-PCR sample loading diagram
5 Are you sure you are doing it right?
Be sure to wear gloves, wear gloves, wear gloves, and say important things three times.
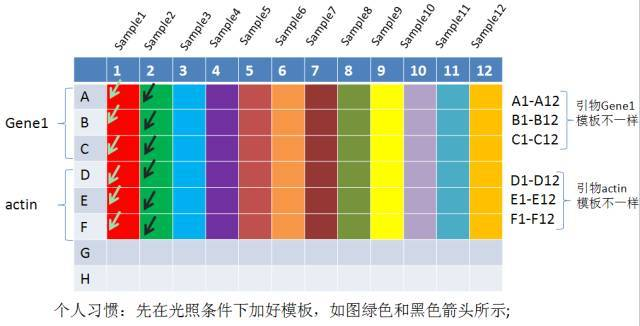
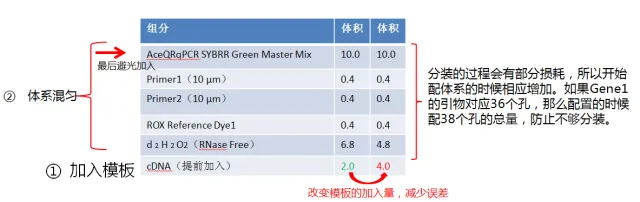
In order to reduce the exposure of SYBR to light, I personally like to add a template first, as shown in the figure below. According to experience, the addition of a small amount of template is likely to cause sampling errors. Therefore, in order to minimize the error caused by adding a small amount of template, I usually double the sample again, and double the amount when adding the sample to reduce the amount of H2O2 added.
Figure. Schematic diagram of qRT-PCR loading
Then configure the qRT-PCR system as follows.
Figure. qRT-PCR system preparation diagram
NOTE: The configuration process needs to be done on ice.
After adding the sample, paste the transparent sealing film. Try not to touch the surface of the transparent sealing film with your hands, just operate from the space on both sides of the film. Because fingerprints may also affect the collection of fluorescent signals. Then use a centrifuge to quickly centrifuge for 10 s at low speed to prevent the sample from hanging on the wall.
Related Products:
Post time: Apr-28-2023