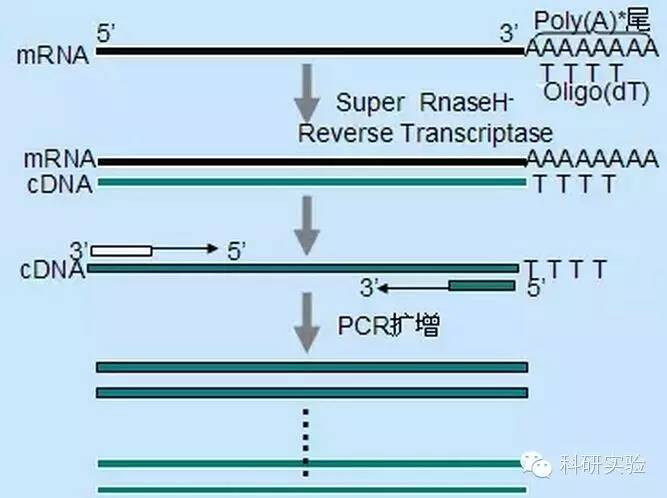
Ⅰ. Increase the sensitivity of the reaction system:
1. Separate high -quality RNA:
Successful cDNA synthesis comes from high -quality RNA. High -quality RNA should ensure at least a total longer and does not contain inhibitors that do not contain recording enzymes, such as EDTA or SDS. The quality of RNA determines the maximum value of the sequence information you can transcribe to the cDNA. The general RNA purification method is a step method of using isoocyanate/acidophenol. In order to prevent the pollution of RNase, the RNA separated from a sample rich in RNase (such as pancreas) requires storage of formaldehyde to save high-quality RNA, which is even more so for long-term storage. The RNA extracted from the rat liver was basically degraded after one week of storage in water, while the RNA extracted from the rat spleen remained stable after three years of storage in water. In addition, transcripts larger than 4kb are more sensitive to trace RNase degradation than small transcripts. In order to increase the stability of the storage RNA sample, the RNA can be dissolved in a methalmamine of ion, and is stored -70 °C. Thylide used to save RNA must not contain a miscellaneous object that degrades RNA. RNA, which is derived from pancreas, can be saved in methalmamine for at least one year. When you are ready to use RNA, you can use the following methods to precipitate RNA: add NaCl to 0.2m and 4 times the volume of ethanol, place room temperature for 3-5 minutes, and 10,000 × g centrifugal for 5 minutes.
2. Use reverse transcriptase without RNaseH activity (RNaseH-):
RNase inhibitors are often added to reverse transcription reactions to increase the length and yield of cDNA synthesis. RNase inhibitor is added in the first chain synthesis reaction in the presence of buffers and reducing agents such as DTT because the pre-cDNA synthesis process denatures the inhibitor, thereby releasing bound RNases that degrade RNA. Protein RNase inhibitor only prevents the degradation of RNA by RNase A, B, C, and do not prevent RNases on the skin, so care should be taken not to introduce RNases from the fingers despite the use of these inhibitors.
Reverse transcriptase catalyzes the conversion of RNA into cDNA. Both M-MLV and AMV have endogenous RNaseH activity in addition to their own polymerase activity. RNaseH activity competes with polymerase activity for heterozygous strands formed between RNA templates and DNA primers or cDNA extension strands, and degrades RNA: RNA strands in DNA complexes. RNA templates degraded by RNaseH activity can no longer be used as effective substrates for cDNA synthesis, reducing the yield and length of cDNA synthesis. Thus eliminating or greatly reducing RNaseH activity of reverse transcriptase would be of great benefit.
SuperScriptⅡ reverse transcriptase, MMLV reverse transcriptase of RNaseH- and thermoScript reverse transcriptase, AMV of RNaseH- yielded more full-length cDNA than MMLV and AMV. RT-PCR sensitivity is affected by the amount of cDNA synthesized. ThermoScript is much more sensitive than AMV. The size of RT-PCR products is limited by the ability of reverse transcriptase to synthesize cDNA, especially when cloning larger Cdnas. Compared with MMLV, SuperScripⅡ significantly increased the yield of long RT-PCR products. RNaseH-’s reverse transcriptase also increases thermal stability, so the reaction can be carried out at temperatures higher than normal of 37-42℃. Under the suggested synthesis conditions, oligo(dT) primers and 10μCi [alpha-p]dCTP were used. The total production of the first chain was calculated using the TCA precipitation method. Full-length cDNA was analyzed using size-sorted strip removal and counting in an alkaline agarose gel.
3. Increase the heat preservation temperature of reverse transcription:
Higher holding temperature helps to open the secondary structure of RNA and increase the yield of the reaction. For most RNA templates, holding the RNA and primer at 65°C without buffer or salt and then cooling them quickly on ice eliminates most secondary structures and allows the primers to bind. However, some templates still have secondary structure, even after thermal denaturation. Amplification of these difficult templates can be performed using ThermoScript reverse transcriptase and by placing the reverse transcriptase reaction at higher temperatures to improve amplification. Higher holding temperatures can also increase specificity, especially when cDNA synthesis is performed using gene-specific primers (GSPS) (see Chapter 3). If using GSP, make sure that the Tm value of the primer is the same as the expected holding temperature. Do not use oligo(dT) and random primers above 60℃. Random primers need to be held at 25℃ for 10 minutes before increasing to 60℃. In addition to using higher reverse transcription temperatures, specificity can be improved by directly transferring the RNA/ primer mixture from the 65℃ denaturing temperature to the reverse transcription holding temperature and adding a preheated 2× reaction mixture (cDNA thermal initiation synthesis). This approach helps prevent the intermolecular base pairing that occurs at lower temperatures. Using a PCR instrument simplifies the many temperature switches required for RT-PCR.
Tth heat stabilized polymerase acts as DNA polymerase in the presence of Mg2+ and RNA polymerase in the presence of Mn2+. It can hold heat at up to 65℃. However, the presence of Mn2+ during PCR reduces fidelity, which makes Tth polymerase less suitable for high precision amplification, such as cDNA cloning. In addition, Tth is less efficient at reverse transcription, which reduces sensitivity, and since a single enzyme can perform reverse transcription and PCR, control reactions without reverse transcription cannot be used to distinguish amplified products of cDNA from those of contaminated genomic DNA.
4. Additive that promotes reverse transcription:
The addition of additives, including glycerin and DMSO, to the first chain synthesis reaction can reduce the stability of the nucleic acid double strand and unwind the RNA secondary structure. Up to 20% glycerin or 10% DMSO can be added without affecting the activity of SuperScriptⅡ or MMLV. AMV can also tolerate up to 20% glycerol without reducing activity. To maximize the sensitivity of RT-PCR in SuperScriptⅡ reverse transcription reaction, 10% glycerol can be added and insulated at 45℃. If 1/10 of the retrotranscription-reaction product is added to the PCR, the concentration of glycerol in the amplification reaction is 0.4%, which is not enough to inhibit PCR.
5. RNaseH processing:
Sensitivity can be improved by treating cDNA synthesis reactions with RNaseH prior to PCR. For some templates, it is thought that the RNA in the cDNA synthesis reaction prevents the binding of amplified products, in which case the RNaseH treatment can increase sensitivity. Generally, RNaseH treatment is required for the amplification of a relatively long full-length cDNA target template, such as tuberous scherosisⅡ with low copy. For this difficult template, RNaseH enhanced the signal generated by the cDNA synthesized by SuperScriptⅡ or AMV. For most RT-PCR reactions, the RNaseH treatment is optional because the 95℃ insulated PCR denaturation step typically hydrolyzes the RNA from the RNA: DNA complex.
6. Improved methods for detecting small amounts of RNA:
RT-PCR is particularly challenging when only small amounts of RNA are available. The addition of glycogen as a carrier during RNA separation helps to increase the yield of small samples. An RNase-free glycogen can be added at the same time as Trizol. Glycogen is water-soluble and can remain in the water phase with RNA to assist in subsequent precipitation. The recommended concentration of RNase-free glycogen is 250μg/ml for samples less than 50mg of tissue or 106 cultured cells.
The addition of acetylated BSA to reverse transcription reactions using SuperScriptⅡ can increase the sensitivity, and for small amounts of RNA, reducing the amount of SuperScriptⅡ and adding 40 units of RnaseOut nuclease inhibitor can improve the detection level. If glycogen is used in RNA separation, the addition of BSA or RNase inhibitors to reverse transcription reactions using SuperScriptⅡ is still recommended.
Ⅱ. Increase the specificity of RT-PCR
1. cNDA synthesis:
Three different methods can be used to initiate first strand cDNA synthesis, and the relative specificity of each method affects the amount and type of cDNA synthesized.
Random primer method is the least specific of the three methods. Primers are annealed at multiple sites throughout the transcript to produce short, partial length cDNA. This method is often used to obtain 5′ terminal sequences and cDNA from RNA templates with secondary structural regions or with terminating sites that reverse transcriptase cannot replicate. To obtain the longest cDNA, the ratio of primers to RNA in each RNA sample needs to be determined empirically. The initial concentration of random primers ranges from 50 to 250ng per 20μl reaction system. Because the cDNA synthesized from total RNA using random primers is mainly ribosomal RNA, poly(A)+RNA is generally selected as the template.
Oligo(dT) initiation is more specific than random primers. It hybridizes with the poly(A) tail found at the 3′ end of mRNA in most eukaryotic cells. Because poly(A)+RNA is roughly 1% to 2% of total RNA, the amount and complexity of cDNA is much less than if random primers were used. Because of its high specificity, oligo(dT) generally does not require optimization for RNA to primer ratio and poly(A)+ selection. It is recommended to use 0.5μg oligo(dT) per 20μl reaction system. oligo(dT)12-18 is suitable for most RT-PCR. ThermoScript RT-PCR System provides oligo(dT)20 because of its good thermal stability and is suitable for higher holding temperatures.
Gene-specific primers (GSP) are the best specific primers for the reverse transcription step. GSP is an antisense oligonucleoside that can specifically hybridize with RNA destination sequences, rather than anneal all Rnas like random primers or oligo(dT). The rules used to design PCR primers also apply to the design of reverse transcription reaction GSP. GSP can be the same sequence as the amplification primer annealed at the end of mRNA3′, or GSP can be designed to be annealed downstream with the reverse amplification primer. For some amplified objects, it is necessary to design more than one antisense primer for successful RT-PCR because the secondary structure of the target RNA may prevent the primer from binding. It is suggested to use 1pmol antisense GSP in the first chain synthesis reaction system of 20μl.
2. Increase the heat preservation temperature of reverse transcription:
In order to take full advantage of GSP specificity, reverse transcriptase with high thermal stability should be used. Heat-stable reverse transcriptase can be insulated at higher temperatures to increase reaction rigor. For example, if a GSP is annealed at 55°C, then the specificity of GSP is not fully utilized if reverse transcription is performed at 37°C with low rigor using AMV or M-MLV. However, SuperScripⅡ and ThermoScript can react at 50℃ or higher, which eliminates nonspecific products produced at lower temperatures. For maximum specificity, the RNA/ primer mixture can be transferred directly from the 65℃ denaturation temperature to the reverse transcription holding temperature with the addition of a preheated 2 x reaction mixture (thermal initiation of cDNA synthesis). This helps prevent base pairing between molecules at low temperatures. Using a PCR instrument simplifies the many temperature transitions required for RT-PCR.
3. Reduce genomic DNA contamination:
One potential difficulty with RT-PCR is that RNA contaminate genomic DNA. The use of better RNA separation methods, such as Trizol Reagent, reduces genomic DNA contamination in RNA preparations. To avoid products produced from genomic DNA, the RNA can be treated with amplification grade DnasⅠ to remove contaminated DNA prior to reverse transcription. The samples were kept at 65℃ in 2.0mM EDTA for 10 min to terminate DNaseⅠ digestion. EDTA chelates magnesium ions to prevent the magnesium ion dependent RNA hydrolysis that occurs at high temperatures.
In order to separate amplified cDNA from the genome DNA amplification product, primers that anneal separately with the separated exon can be designed. PCR products derived from cDNA will be shorter than those derived from contaminated genomic DNA. A controlled experiment without reverse transcription is also performed on each RNA template to determine whether a given fragment is from genomic DNA or cDNA. PCR products obtained in the absence of reverse transcription are derived from the genome.
Related Product
-The one-step kit enables reverse transcription and PCR to be carried out in the same tube. It only needs to add template RNA, specific PCR primers and RNase-Free ddH2O.
-Real-time quantitative analysis of RNA can be carried out quickly and accurately.
-The kit uses a unique Foregene reverse transcription reagent and Foregene HotStar Taq DNA Polymerase combined with a unique reaction system to effectively improve the amplification efficiency and specificity of the reaction.
-The optimized reaction system makes the reaction have higher detection sensitivity, stronger thermal stability, and better tolerance.
RT Easy II(With GDNase) Master Premix For First-Strand CDNA Synthesis For Real Time PCR With GDNase
-Efficient ability to remove gDNA, which can remove gDNA in the template within 2 minutes.
-Efficient reverse transcription system, it only takes 15 minutes to complete the synthesis of the first strand cDNA.
-Complex templates: templates with high GC content and complex secondary structure can also be reversed with high efficiency.
-High-sensitivity reverse transcription system, pg-level templates can also get high-quality cDNA.
-The reverse transcription system has high thermal stability, the optimal reaction temperature is 42℃, and it still has good reverse transcription performance at 50℃.
Post time: Mar-07-2023