The three letters SNP are ubiquitous in the study of population genetics. Regardless of human disease research, crop trait positioning, animal evolution and molecular ecology, SNPs are needed as the basis. However, if you don’t have a deep understanding of modern genetics based on high-throughput sequencing, and facing these three letters, it seems "the most familiar stranger", then you can't carry out the subsequent research. So before embarking on follow-up research, let's take a look at what SNP is.
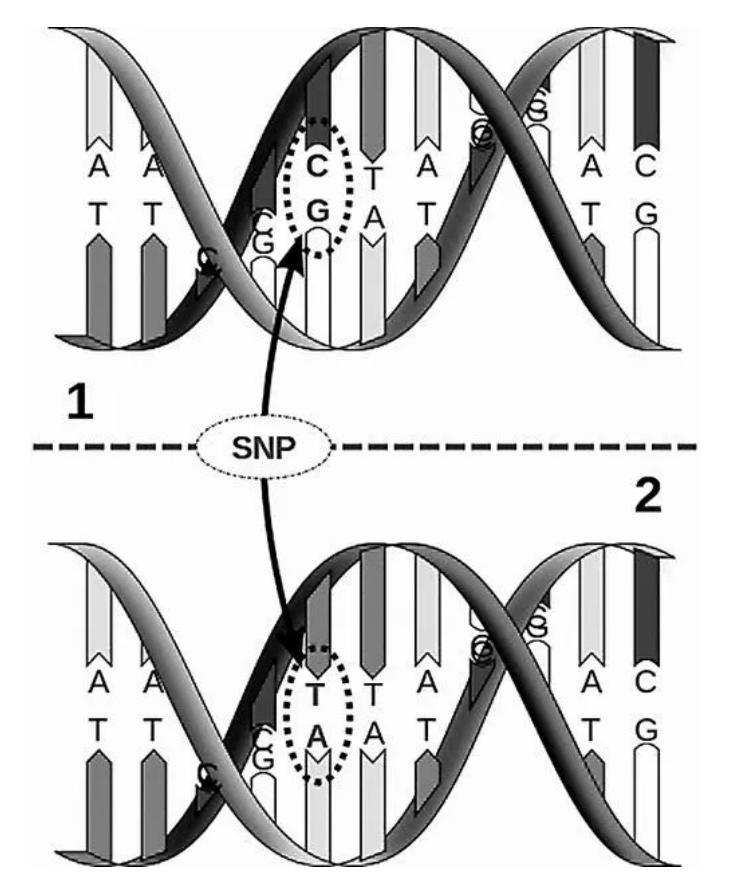
SNP (single nucleotide polymorphism), we can see from its full English name, it refers to single nucleotide variation or polymorphism. It also has a separate name, called SNV (single nucleotide variation). In some human studies, only those with a population frequency higher than 1% are called SNPs, but broadly speaking, the two can be mixed. So we can say that SNP, single nucleotide polymorphism, refers to the mutation in which one nucleotide in the genome is replaced by another nucleotide. For example, in the figure below, an AT base pair is replaced with a GC base pair, which is a SNP site.
Picture
However, whether it is a "single nucleotide polymorphism" or a "single nucleotide variation", it is relatively speaking, so SNP data needs genome resequencing as the basis, that is, the sequencing data is sequenced after the individual's genome is sequenced. Compared to the genome, the site that differs from the genome is detected as a SNP site.
In terms of SNPs on vegetables, the Plant Direct PCR kit can be used for fast detection.
In terms of mutation types, SNP includes transition and transversion. Transition refers to the replacement of purines with purines or pyrimidines with pyrimidines. Transversion refers to the mutual substitution between purines and pyrimidines. The frequency of occurrence will be different, and the probability of transition will be higher than that of transversion.
In terms of where the SNP occurs, different SNPs will have different effects on the genome. SNPs that occur in the intergenic region, that is, the region between genes on the genome, may not affect the function of the genome, and mutations in the intron or the upstream promoter region of the gene may have a certain impact on the gene; The mutations that occur in the exon regions of genes, depending on whether they cause changes in the encoded amino acids, have different effects on gene functions. (Of course, even if two SNPs cause differences in amino acids, they have different effects on the structure of the protein, and ultimately the effects on the biological phenotype may be quite different).
However, the number of SNPs that occur at gene locations is usually significantly less than that of non-gene locations, because a SNP that affects the function of a gene usually has a negative impact on the survival of the individual, resulting in the individual carrying this SNP in the group Among them was eliminated.
Of course, for diploid organisms, chromosomes exist in pairs, but it is impossible for a pair of chromosomes to be exactly the same for every base. Therefore, some SNPs will also appear heterozygous, that is, there are two bases at this position on the chromosome. In a group, the SNP genotypes of different individuals are aggregated together, which becomes the basis for most subsequent analyses. In combination with traits, it can be judged whether SNP as a molecular marker is linked to traits, the QTL (quantitative trait locus) of the trait can be judged, and GWAS (genome-wide association study) or genetic map construction can be performed; SNP can be used as a molecular marker Judge the evolutionary relationship between individuals; you can screen functional SNPs and study disease-related mutations; you can use SNP allele frequency changes or heterozygous rates and other indicators to determine the selected regions on the genome... etc., combined with the present With the development of high-throughput sequencing, hundreds of thousands or more SNP sites can be obtained from a set of sequencing data. It can be said that SNP has now become the cornerstone of population genetic research.
Of course, the changes in the bases in the genome are not always the replacement of one base with another (although this is the most common one). It is also possible that one or a few bases are missing, or two bases. Several other bases were inserted in the middle. This small range of insertions and deletions is collectively called InDel (insertion and deletion), which specifically refers to the insertion and deletion of short fragments (one or several bases). InDel that occurs at the location of a gene may also have an impact on the function of the gene, so sometimes InDel may also play an important role in research. But overall, the status of SNP as the cornerstone of population genetics is still unshakable.
Post time: Aug-27-2021