In qPCR experiments, primer design is also a very important link. Whether the primers are suitable or not is closely related to whether the amplification efficiency reaches the standard, whether the amplified products are specific, and whether the experimental results are available.
So how to make qPCR primer specificity better? High amplification efficiency?
Today, we will take you to design qPCR primers together, and let qPCR primer design become an efficient lore skill in experiments.
When designing qPCR primers, usually pay attention to the following points: primers should be designed across introns as much as possible, the product length should be 100-300 bp, the Tm value should be as close as possible to 60°C, and the upstream and downstream primers should be as close as possible, and the end of the primer should be G or C, etc. wait.
1. Design of primers spanning introns
When designing qPCR primers, choosing primers designed across introns can prevent the gDNA template from being amplified, and the products are all derived from the amplification of cDNA, thus eliminating the influence of gDNA contamination.
2. Primer length
The primer length is generally between 18-30 nt, and the length of the amplification product should be controlled between 100-300 bp as much as possible.
If the primer is too short, it will lead to non-specific amplification, and if it is too long, it will easily form secondary structure (such as hairpin structure). If the amplification product is too long, it is not suitable for the reaction of polymerase, which will affect the efficiency of PCR amplification.
3. GC content and Tm value
The GC content of primers should be controlled between 40% and 60%. If it is too high or too low, it is not conducive to initiating the reaction. The GC content of the forward and reverse primers should be close to the same in order to obtain the same Tm value and annealing temperature.
The Tm value should be between 55-65°C as far as possible, generally around 60°C, and the Tm value of the upstream and downstream should be as close as possible, preferably no more than 4°C.
4. Avoid selecting A at the 3′ end of the primer
When the 3′ end of the primer is mismatched, there are great differences in the synthesis efficiency of different bases. When the last base is A, it can also initiate chain synthesis even in the case of mismatching, and when the last base is T When , the efficiency of mismatch induction is greatly reduced. Therefore, try to avoid choosing A at the 3′ end of the primer, and it is better to choose T.
If it is a probe primer, the 5′ end of the probe cannot be G, because even when a single G base is connected to the FAM fluorescent reporter group, G can also quench the fluorescent signal emitted by the FAM group, resulting in false negative results. Appear.
5. Base distribution
The distribution of the four bases in the primer is preferably random, avoiding more than 3 consecutive G or C at the 3′ end, and more than 3 consecutive G or C are easy to generate pairing in the GC-rich sequence region.
6. The primer design region should avoid complex secondary structures.
The secondary structure formed by the single strand of the amplification product will affect the smooth progress of PCR. By predicting whether there is a secondary structure in the target sequence in advance, try to avoid this region in the design of primers.
7. The primers themselves and between the primers should try to avoid consecutive complementary bases.
There can be no consecutive 4 base complementarity between the primer itself and the primer. The primer itself should not have a complementary sequence, otherwise it will fold itself to form a hairpin structure, which will affect the annealing combination of the primer and the template.
Complementary sequences cannot exist between upstream and downstream primers. Complementarity between primers will produce primer dimers, which will reduce PCR efficiency and even affect quantitative accuracy. If the primer-dimer and hairpin structures are unavoidable, the △G value should not be too high (should be less than 4.5 kcal/mol).
8. The primers amplify the target specific product.
The ultimate goal of qPCR detection is to understand the abundance of the target gene. If non-specific amplification occurs, the quantification will be inaccurate. Therefore, after the primers are designed, they need to be tested by BLAST, and the specificity of the products is compared in the sequence database.
Next, we take the human GAS6 (Growth arrest specific 6) gene as an example to design qPCR primers.
01 query gene
Homo GAS6 through NCBI . Here, we should pay attention to comparing the gene name and species to ensure that they are consistent.
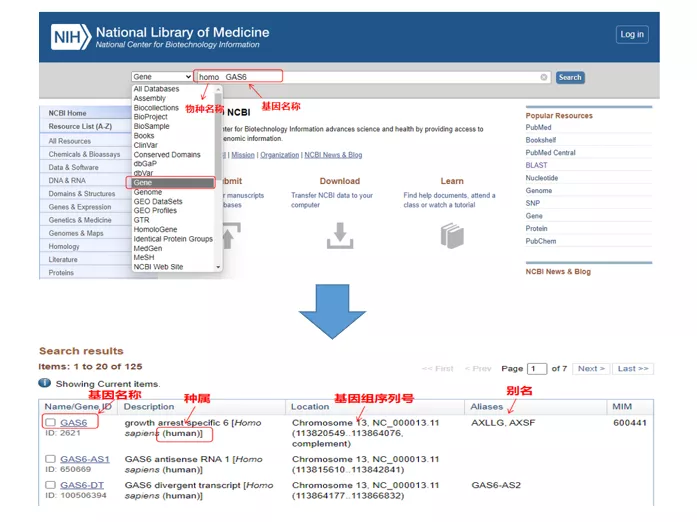 02 Find the gene sequence
02 Find the gene sequence
(1) If the target sequence is genomic DNA, select the first one, which is the genomic DNA sequence of the gene.
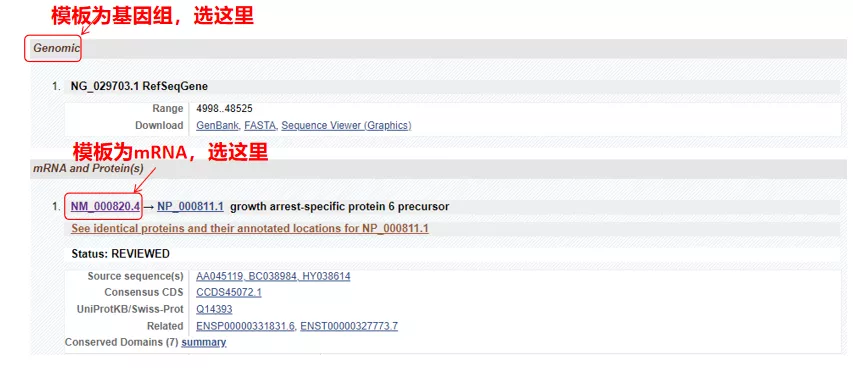 (2) If the target sequence is mRNA, select the second one. After entering, click “CDS” in the table below. The brown background sequence is the coding sequence of the gene.
(2) If the target sequence is mRNA, select the second one. After entering, click “CDS” in the table below. The brown background sequence is the coding sequence of the gene.
 03 Design primers
03 Design primers
Enter the Primer-BLAST interface
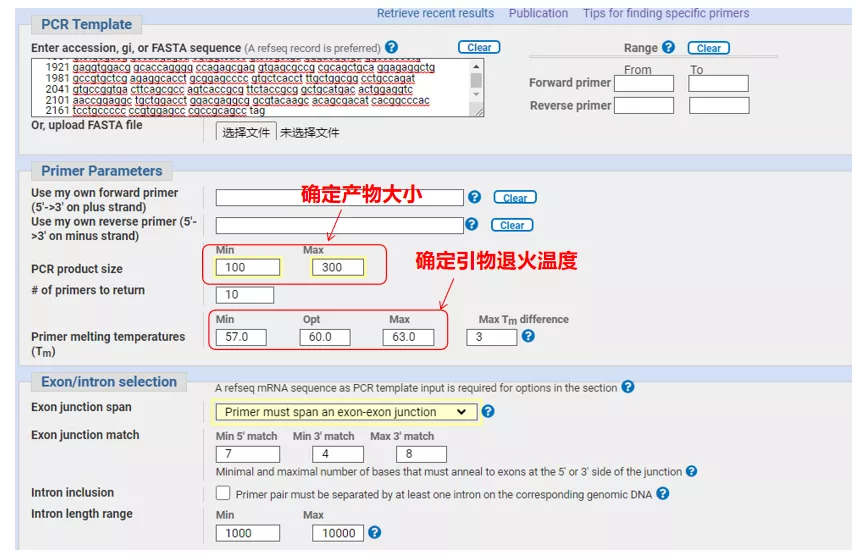
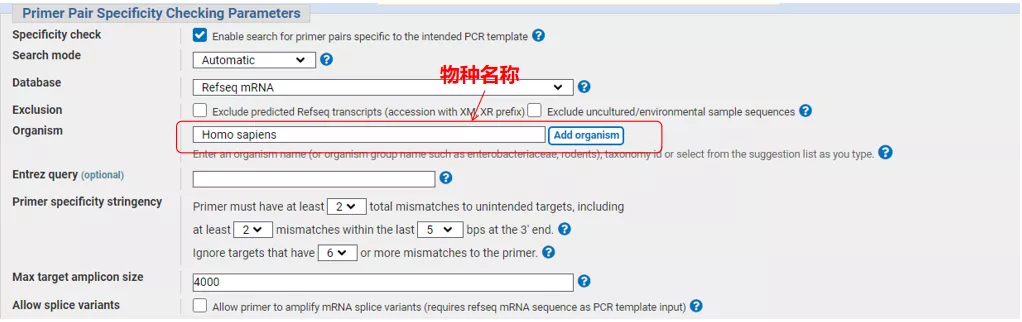
 Enter the gene sequence number or the sequence in Fasta format on the upper left, and fill in the relevant parameters.
Enter the gene sequence number or the sequence in Fasta format on the upper left, and fill in the relevant parameters.
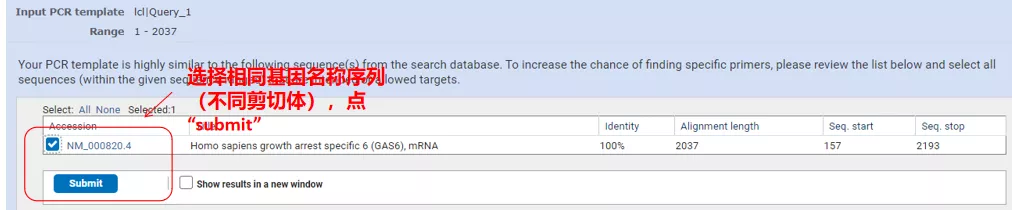
Click “Get primers” and NCBI will pop up to tell you that such parameter selection will be amplified to other splicing variants. We can check the different splicing variants and submit them to get the appropriate primer pair (as shown in the figure below ). This process may take tens of seconds to run.
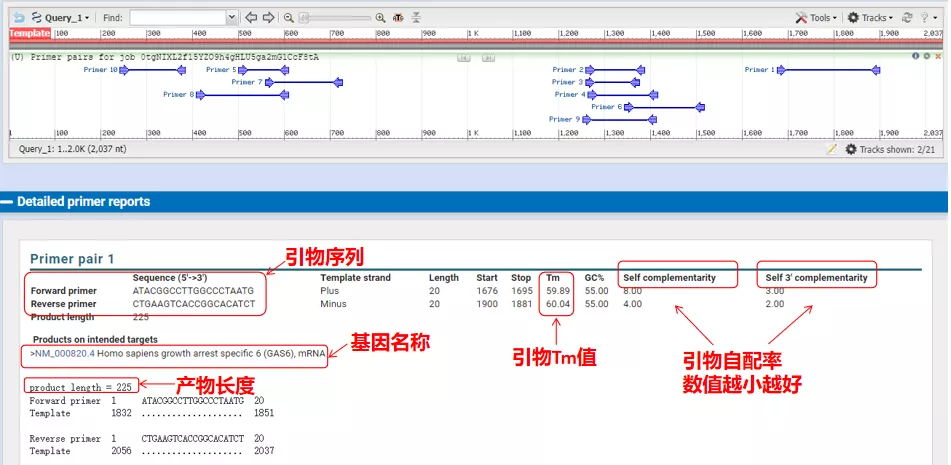 The annealing temperatures of these primer pairs are all around 60°C. According to the purpose of the experiment, choose primers with moderate length, good specificity and less self-complementation of the primers for the experiment, and the success rate is quite high!
The annealing temperatures of these primer pairs are all around 60°C. According to the purpose of the experiment, choose primers with moderate length, good specificity and less self-complementation of the primers for the experiment, and the success rate is quite high!
04Primer specificity verification
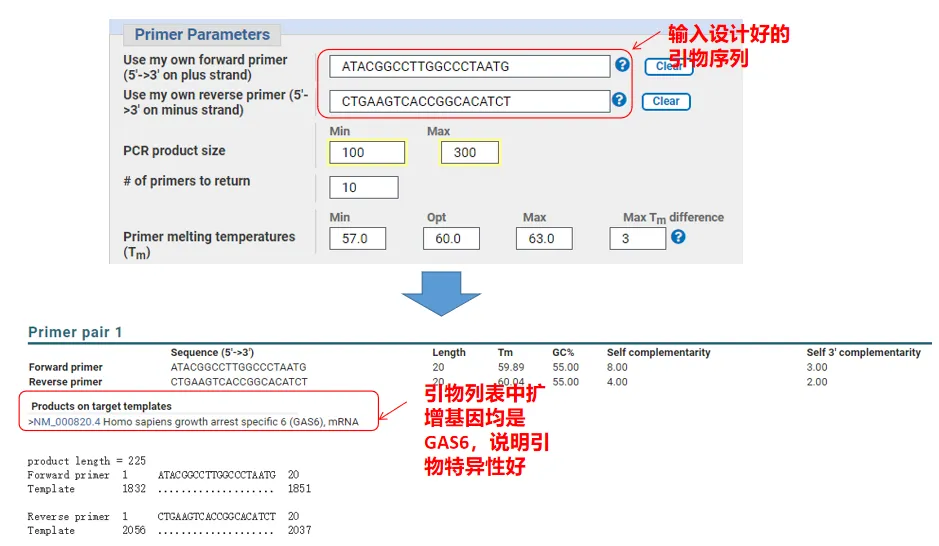
In fact, in addition to designing primers, Primer-Blast can also evaluate the primers we designed ourselves. Return to the primer design page, enter the upstream and downstream primers we designed, and other parameters will not be adjusted. After submitting, you can see whether the pair of primers also exist on other genes. If all of them are displayed on the gene we want to amplify , indicating that the specificity of this pair of primers is great! (For example, this is the only result of the primer query!)
05 Primer quality judgment
What kind of primer is the “perfect” primer that combines “amplification efficiency up to standard”, “amplified product characteristics”, and “reliable experimental results”?
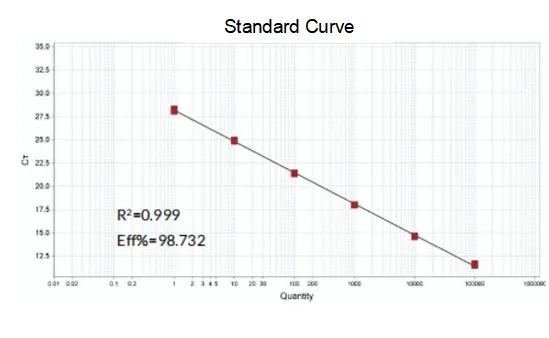 Amplification efficiency
Amplification efficiency
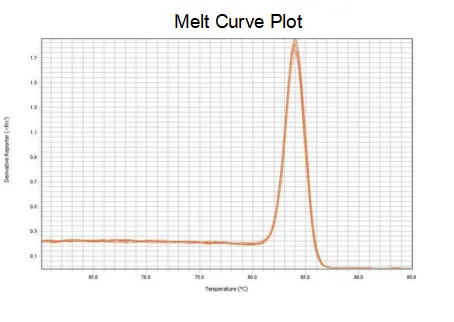 melting curve
melting curve
The amplification efficiency of the primers reaches 90%-110%, which means that the amplification efficiency is good, and the melting curve has a single peak and usually Tm>80°C, which means that the amplification specificity is good.
Related Products:
Real Time PCR Easy–SYBR GREEN I
Real Time PCR Easy-Taqman
Post time: Feb-10-2023








