Starting material: RNA
Quantitative reverse transcription PCR (RT-qPCR) is an experimental method used in PCR experiments using RNA as the starting material. In this method, total RNA or messenger RNA (mRNA) is first transcribed into complementary DNA (cDNA) by reverse transcriptase. Subsequently, a qPCR reaction was performed using the cDNA as a template. RT-qPCR has been used in a variety of molecular biology applications, including gene expression analysis, RNA interference validation, microarray validation, pathogen detection, genetic testing, and disease research.
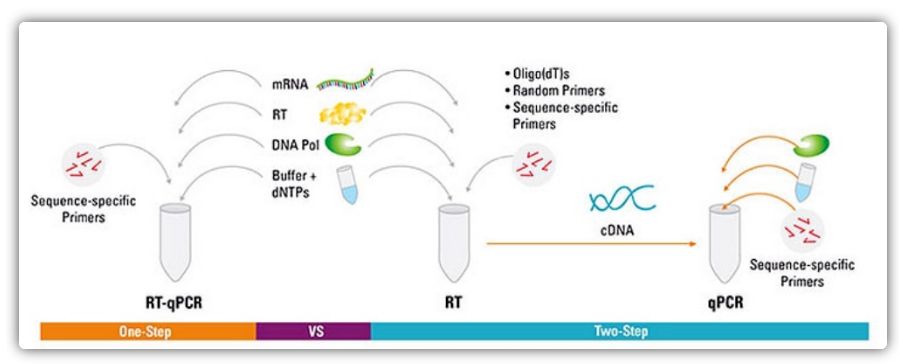
One-step and two-step methods for RT-qPCR
RT-qPCR can be accomplished by a one-step or two-step method. One-step RT-qPCR combines reverse transcription and PCR amplification, allowing reverse transcriptase and DNA polymerase to complete the reaction in the same tube under the same buffer conditions. One-step RT-qPCR only requires the use of sequence-specific primers. In two-step RT-qPCR, reverse transcription and PCR amplification are performed in two tubes, using different optimized buffers, reaction conditions, and primer design strategies.
|
Advantage |
Disadvantage |
|
| One Step | This method has less experimental error as both reactions are done in one tube
Fewer pipetting steps reduce the risk of contamination
Suitable for high-throughput amplification/screening, fast and reproducible |
Two-step reactions cannot be optimized separately
Since the reaction conditions are compromised by combining the two-step reaction, the sensitivity is not as good as that of the two-step method
The number of targets detected by a single sample is small |
| Two Steps | Ability to create stable cDNA libraries that can be stored for long periods of time and used in multiple reactions
Target genes and reference genes can be amplified from the same cDNA library without the need for multiple cDNA libraries
Reaction buffers and reaction conditions that enable optimization of single reaction runs
Flexible selection of trigger conditions |
Using multiple tubes, and more pipetting steps increases the risk of DNA contamination,
and time consuming.
Requires more optimization than the one-step method |
Related products:
RT-qPCR Easyᵀᴹ (One Step)-SYBR Green I
RT-qPCR Easyᵀᴹ (One Step)-Taqman
RT Easyᵀᴹ I Master Premix For First-Strand CDNA Synthesis
Real Time PCR Easyᵀᴹ-SYBR Green I Kit
Selection of total RNA and mRNA
When designing an RT-qPCR experiment, it is important to decide whether to use total RNA or purified mRNA as a template for reverse transcription. Although mRNA may be able to provide slightly higher sensitivity, total RNA is still frequently used. The reason for this is that total RNA has a more important advantage as a starting material than mRNA. First, the process requires fewer purification steps, which ensures better quantitative recovery of template and better normalization of results to starting cell numbers. Second, it avoids the mRNA enrichment step, which can avoid the possibility of skewed results due to different recoveries of different mRNAs. Overall, since in most applications the relative quantification of the target gene is more important than the absolute sensitivity of the detection, total RNA is more suitable in most cases.
Reverse transcription primer
In the two-step method, three different methods can be used to prime the cDNA reaction: oligo(dT) primers, random primers, or sequence-specific primers. Typically, oligo(dT) primers and random primers are used in combination. These primers anneal to the template mRNA strand and provide reverse transcriptase with a starting point for synthesis.
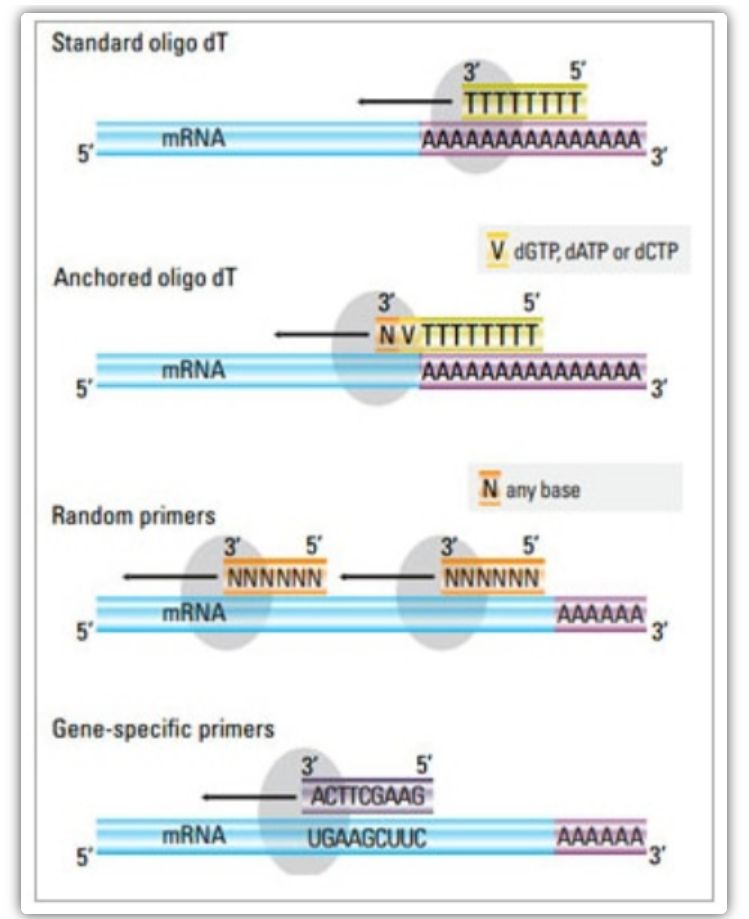
| Primer selection | Structure and function | Advantage | Disadvantage |
| Oligo(dT) primer (or anchored oligo(dT) primer) | Extended annealing to thymine residues at the poly(A) tail of mRNA; anchor oligo(dT) primer contains a G, C, or A at the 3′ end (anchor site) | Synthesis of full-length cDNA from poly(A)-tailed mRNA
Applicable when less starting material is available
Anchoring site ensures that the oligo(dT) primer binds to the 5′ poly(A) tail of the mRNA |
Only suitable for amplifying genes with poly(A) tails
Obtain cDNA truncated from the priming site*2 in poly(A)
Biased to bind to the 3′ end*
*This possibility is minimized if anchored oligo(dT) primers are used |
| random primer
|
6 to 9 bases in length, which can anneal to multiple sites during RNA transcription | Anneal to all RNAs (tRNA, rRNA, and mRNA)
Suitable for transcripts with significant secondary structure, or when less starting material is available
High cDNA Yield |
cDNA is reverse transcribed from all RNA, which is usually not desired and may dilute the signal of the target mRNA
get truncated cDNA |
| sequence-specific primers | Custom primers targeting specific mRNA sequences | specific cDNA library
Improve sensitivity
Using reverse qPCR primers |
Only limited to the synthesis of a single target gene |
Reverse transcriptase
Reverse transcriptase is an enzyme that uses RNA to synthesize DNA. Some reverse transcriptases have RNase activity and can degrade RNA strands in RNA-DNA hybrid strands after transcription. If it does not have RNase enzymatic activity, RNaseH can be added for higher qPCR efficiency. Commonly used enzymes include Moloney murine leukemia virus reverse transcriptase and avian myeloblastoma virus reverse transcriptase. For RT-qPCR, it is ideal to choose a reverse transcriptase with higher thermostability, so that cDNA synthesis can be performed at higher temperatures, ensuring successful transcription of RNAs with higher secondary structure, while maintaining their full activity throughout the reaction, resulting in higher cDNA yields.
Related products:
Foreasy M-MLV Reverse Transcriptase
RNase H activity of reverse transcriptase
RNaseH is able to degrade RNA strands from RNA-DNA duplexes, allowing efficient synthesis of double-stranded DNA. However, when using long mRNA as a template, the RNA may be degraded prematurely, resulting in truncated cDNA. Therefore, it is often beneficial to minimize RNaseH activity during cDNA cloning if synthesis of long transcripts is desired. In contrast, reverse transcriptases with RNase H activity are often beneficial for qPCR applications because they enhance the melting of RNA-DNA duplexes during the first cycle of PCR.
Primer design
PCR primers used for the qPCR step in RT-qPCR should ideally be designed to span an exon-exon junction, where an amplification primer could potentially span an actual exon-intron boundary. Since intron-containing genomic DNA sequences are not amplified, this design reduces the risk of false positives amplified from contaminating genomic DNA.
If primers cannot be designed to separate exons or exon-exon boundaries, it may be necessary to treat RNA samples with RNase-free DNase I or dsDNase to remove genomic DNA contamination.
RT-qPCR control
A reverse transcription negative control (-RT control) should be included in all RT-qPCR experiments to detect DNA contamination (such as genomic DNA or PCR products from previous reactions). This control contains all reaction components except reverse transcriptase. Since reverse transcription does not occur with this control, if PCR amplification is observed, contamination from DNA is most likely.
Post time: Aug-02-2022








